Difference between revisions of "Influenza A"
Techsensus (talk | contribs) |
Techsensus (talk | contribs) (→Safety) |
||
| (38 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
[[File:Influenzastructure.png|thumb|360px|Structure of the influenza virus.<ref name="Arti1">Host Protective Immune Responses against Influenza A Virus Infection, MDPI, 2020, Hi Eun Jung, Heung Kyu Lee, https://www.mdpi.com/1999-4915/12/5/504/htm</ref>]] | [[File:Influenzastructure.png|thumb|360px|Structure of the influenza virus.<ref name="Arti1">Host Protective Immune Responses against Influenza A Virus Infection, MDPI, 2020, Hi Eun Jung, Heung Kyu Lee, https://www.mdpi.com/1999-4915/12/5/504/htm</ref>]] | ||
| − | == General | + | == General information== |
| − | The theme of SensUs 2021 is acute respiratory viruses. The current Covid-19 pandemic has made it apparent that large virus outbreaks can cause immense harm to human health and can disrupt society as a whole. | + | The theme of SensUs 2021 is acute respiratory viruses. The current Covid-19 pandemic has made it apparent that large virus outbreaks can cause immense harm to human health and can disrupt society as a whole. One of the most common respiratory viruses is influenza A. Therefore, the influenza virus serves as an interesting research topic for this year’s Competition. |
| − | The influenza A virus is known to cause influenza in birds and some mammals, like humans. | + | The influenza A virus is known to cause influenza in birds and some mammals, like humans. Several subtypes of the influenza A virus have been isolated from wild birds. Occasionally, viruses can be transmitted from wild birds to domestic animals, like chickens or pigs. This may give rise to human influenza.<ref name="Arti2">Transmission of Avian Influenza A Viruses Between Animals and People, CDC, 2015, https://www.cdc.gov/flu/avianflu/virus-transmission.htm</ref> |
| − | Influenza A viruses are | + | Influenza A viruses are negative-sense, single-stranded RNA viruses. Subtypes of influenza A are characterized by proteins on the outermost membrane of the virus, called hemagglutinin (H or HA) and neuraminidase (N or NA). H and N are the antigens of the virus, and play an important role in the interaction between the host’s immunological response and the virus. Recently, researchers have reported the discovery of an antibody that is generally effective against all subtypes of the influenza A virus.<ref name="Arti3">Super antibody' fights off flu, BBC, 2011, James Gallagher, https://www.bbc.com/news/health-14324901</ref> Hemagglutinin will serve as this year's biomarker. In infected patients, bound hemagglutinin indicates the presence of the influenza virus. In the Testing Event, unbound hemagglutinin proteins will be used to mimic the immunologic response of the virus. |
| − | The subtype which will be used in SensUs 2021 is H1N1. Historically, H1N1 has been responsible for most deaths due to influenza. It is a popular influenza strain for research purposes.<ref name="Arti4">Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection, Research Gate, 2020, Sun, Honglei & Xiao, Yihong & Liu, Jiyu & Wang, Dayan & Li, Fangtao & Wang, Chenxi & Li, Chong & Zhu, Junda & Song, Jingwei & Sun, Haoran & Zhimin, Jiang & Liu, Litao & Zhang, Xin & Wei, Kai & Dongjun, Hou & Pu, Juan & Sun, Yipeng & Tong, Qi & Bi, Yuhai & Liu, Jinhua https://www.researchgate.net/publication/342555087_Prevalent_Eurasian_avian-like_H1N1_swine_influenza_virus_with_2009_pandemic_viral_genes_facilitating_human_infection</ref><ref name="Arti5">Comparison of Hospitalized Patients With ARDS Caused by COVID-19 and H1N1, CHEST, Xiao Tang, Rong-Hui Du, Rui Wang, Tan-Ze Cao, Lu-Lu Guan, Cheng-Qing Yang, Qi Zhu, Ming Hu, Xu-Yan Li, Ying Li, Li-Rong Liang, Zhao-Hui Tong, Bing Sun, Peng Peng, Huan-Zhong Shi, 2020, https://www.sciencedirect.com/science/article/pii/S0012369220305584</ref><ref name="Arti6">Landscape of coordinated immune responses to H1N1 challenge in humans, Journal of Clinical Investigation, 2020, Zainab Rahil, Rebecca Leylek, Christian M. Schürch, Han Chen, Zach Bjornson-Hooper, Shannon R. Christensen, Pier Federico Gherardini, Salil S. Bhate, Matthew H. Spitzer, Gabriela K. Fragiadakis, Nilanjan Mukherjee, Nelson Kim, Sizun Jiang, Jennifer Yo, Brice Gaudilliere, Melton Affrime, Bonnie Bock, Scott E. Hensley, Juliana Idoyaga, Nima Aghaeepour, Kenneth Kim, Garry P. Nolan, David R. McIlwain https://www.sciencedirect.com/science/article/pii/S0012369220305584</ref> Due to its popularity among researchers, antigens and antibodies are commercially available, making H1N1 suitable as a target for the SensUs | + | The subtype which will be used in SensUs 2021 is H1N1. Historically, H1N1 has been responsible for most deaths due to influenza. It is a popular influenza strain for research purposes.<ref name="Arti4">Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection, Research Gate, 2020, Sun, Honglei & Xiao, Yihong & Liu, Jiyu & Wang, Dayan & Li, Fangtao & Wang, Chenxi & Li, Chong & Zhu, Junda & Song, Jingwei & Sun, Haoran & Zhimin, Jiang & Liu, Litao & Zhang, Xin & Wei, Kai & Dongjun, Hou & Pu, Juan & Sun, Yipeng & Tong, Qi & Bi, Yuhai & Liu, Jinhua https://www.researchgate.net/publication/342555087_Prevalent_Eurasian_avian-like_H1N1_swine_influenza_virus_with_2009_pandemic_viral_genes_facilitating_human_infection</ref><ref name="Arti5">Comparison of Hospitalized Patients With ARDS Caused by COVID-19 and H1N1, CHEST, Xiao Tang, Rong-Hui Du, Rui Wang, Tan-Ze Cao, Lu-Lu Guan, Cheng-Qing Yang, Qi Zhu, Ming Hu, Xu-Yan Li, Ying Li, Li-Rong Liang, Zhao-Hui Tong, Bing Sun, Peng Peng, Huan-Zhong Shi, 2020, https://www.sciencedirect.com/science/article/pii/S0012369220305584</ref><ref name="Arti6">Landscape of coordinated immune responses to H1N1 challenge in humans, Journal of Clinical Investigation, 2020, Zainab Rahil, Rebecca Leylek, Christian M. Schürch, Han Chen, Zach Bjornson-Hooper, Shannon R. Christensen, Pier Federico Gherardini, Salil S. Bhate, Matthew H. Spitzer, Gabriela K. Fragiadakis, Nilanjan Mukherjee, Nelson Kim, Sizun Jiang, Jennifer Yo, Brice Gaudilliere, Melton Affrime, Bonnie Bock, Scott E. Hensley, Juliana Idoyaga, Nima Aghaeepour, Kenneth Kim, Garry P. Nolan, David R. McIlwain https://www.sciencedirect.com/science/article/pii/S0012369220305584</ref> Due to its popularity among researchers, antigens and antibodies are commercially available, making H1N1 suitable as a target for the SensUs Competition. |
Influenza A vaccines for humans have been developed. New versions of the vaccines are developed twice per year for use all over the world, which is necessary due to rapid mutations of the influenza virus. Every year during flu season, a large part of the population is vaccinated in order to protect individuals against the virus. However, due to unforeseen mutations of the virus, it might be possible that in a certain year a vaccine will prove ineffective. In that case large portions of the population would be at risk and a pandemic could occur. The probability of a major influenza A pandemic is estimated to be around 0.5-1% each year.<ref name="Arti7">Pandemic risk: how large are the expected losses?, WHO, 2017, Victoria Y Fan, Dean T Jamisonb & Lawrence H Summers, https://www.who.int/bulletin/volumes/96/2/17-199588.pdf</ref> | Influenza A vaccines for humans have been developed. New versions of the vaccines are developed twice per year for use all over the world, which is necessary due to rapid mutations of the influenza virus. Every year during flu season, a large part of the population is vaccinated in order to protect individuals against the virus. However, due to unforeseen mutations of the virus, it might be possible that in a certain year a vaccine will prove ineffective. In that case large portions of the population would be at risk and a pandemic could occur. The probability of a major influenza A pandemic is estimated to be around 0.5-1% each year.<ref name="Arti7">Pandemic risk: how large are the expected losses?, WHO, 2017, Victoria Y Fan, Dean T Jamisonb & Lawrence H Summers, https://www.who.int/bulletin/volumes/96/2/17-199588.pdf</ref> | ||
==History of influenza (A)== | ==History of influenza (A)== | ||
| − | A lack of data up until 1500 AC complicates the research on influenza before that period.<ref name="Arti8">Internet‐Based Intelligence in Public Health Emergencies, NATO Science for Peace and Security Series - E: Human and Societal Dynamics, 2013 Mordini E., Green M., https://www.iospress.nl/book/internet%E2%80%90based-intelligence-in-public-health-emergencies/</ref> Possibly the first influenza pandemic occurred around 6000 BC in China. The symptoms of human influenza seem to have been clearly described by Hippocrates, roughly 2,400 years ago<ref name="Arti9">2,500-year Evolution of the Term Epidemic, Emerging infectious diseases, 2006, Martin PM, Martin-Granel E, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3373038/</ref> Although the virus seems to have caused epidemics throughout human history, historical data on influenza is difficult to interpret, due to the fact that symptoms of influenza are similar to those found in other respiratory diseases, like | + | A lack of data up until 1500 AC complicates the research on influenza before that period.<ref name="Arti8">Internet‐Based Intelligence in Public Health Emergencies, NATO Science for Peace and Security Series - E: Human and Societal Dynamics, 2013 Mordini E., Green M., https://www.iospress.nl/book/internet%E2%80%90based-intelligence-in-public-health-emergencies/</ref> Possibly the first influenza pandemic occurred around 6000 BC in China. The symptoms of human influenza seem to have been clearly described by Hippocrates, roughly 2,400 years ago<ref name="Arti9">2,500-year Evolution of the Term Epidemic, Emerging infectious diseases, 2006, Martin PM, Martin-Granel E, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3373038/</ref> Although the virus seems to have caused epidemics throughout human history, historical data on influenza is difficult to interpret, due to the fact that symptoms of influenza are similar to those found in other respiratory diseases, like respiratory syncytial virus (RSV). |
| − | The most infamous and lethal outbreak was the 1918 flu pandemic | + | The most infamous and lethal outbreak was the 1918 flu pandemic, which lasted until 1920. The number of deaths is unknown, but estimates range from 17 to 100 million people.<ref name="Arti10">Reassessing the Global Mortality Burden of the 1918 Influenza Pandemic, American journal of epidemiology, 2018, Spreeuwenberg P, Kroneman M, Paget J, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7314216/</ref>. This pandemic has been described as "the greatest medical holocaust in history"<ref name="Arti11"> Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission |
, Pathogens, 2016, Saunders-Hastings PR, Krewski D, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5198166/</ref> and may have killed as many people as the plague (Black Death). This huge death toll was caused by an extremely high infection rate of up to 50% and the severity of the symptoms, suspected to be caused by cytokine storms in which the innate immune system causes an uncontrolled and excessive release of pro-inflammatory signaling molecules called cytokines. | , Pathogens, 2016, Saunders-Hastings PR, Krewski D, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5198166/</ref> and may have killed as many people as the plague (Black Death). This huge death toll was caused by an extremely high infection rate of up to 50% and the severity of the symptoms, suspected to be caused by cytokine storms in which the innate immune system causes an uncontrolled and excessive release of pro-inflammatory signaling molecules called cytokines. | ||
One of the most recent outbreaks of influenza was the 2009 Swine Flu. Similar to the Spanish Flu, it was also of the subtype H1N1. The death toll of the 2009 pandemic is estimated to be around 150,000 to 575,000.<ref name="Arti12">2009 H1N1 Pandemic (H1N1pdm09 virus), centers for disease control and prevention, 2010, https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html</ref> | One of the most recent outbreaks of influenza was the 2009 Swine Flu. Similar to the Spanish Flu, it was also of the subtype H1N1. The death toll of the 2009 pandemic is estimated to be around 150,000 to 575,000.<ref name="Arti12">2009 H1N1 Pandemic (H1N1pdm09 virus), centers for disease control and prevention, 2010, https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html</ref> | ||
| Line 30: | Line 28: | ||
These two proteins are a target of interest for antiviral drugs.<ref name="Arti15">Recent Strategies in the Search for New Anti-Influenza Therapies, Current Drug Targets , 2003, J.C. Wilson, M. von Itzstein, https://www.eurekaselect.com/63785/article</ref> Furthermore, they are also the antigen proteins to which a host antibodies can bind and trigger an immune response. Influenza type A viruses are categorized into different subtypes, or strains, based on which type of these two proteins is present on the surface of the virion. Currently, there are 16 subtypes of HA and 9 subtypes of NA known to exist. The most prevalent form of the different subtypes is H1N1. Single hemagglutinin-neuraminidase proteins, in which both HA and NA are found in a single protein, also exist. However, these will not be used in SensUs 2021. | These two proteins are a target of interest for antiviral drugs.<ref name="Arti15">Recent Strategies in the Search for New Anti-Influenza Therapies, Current Drug Targets , 2003, J.C. Wilson, M. von Itzstein, https://www.eurekaselect.com/63785/article</ref> Furthermore, they are also the antigen proteins to which a host antibodies can bind and trigger an immune response. Influenza type A viruses are categorized into different subtypes, or strains, based on which type of these two proteins is present on the surface of the virion. Currently, there are 16 subtypes of HA and 9 subtypes of NA known to exist. The most prevalent form of the different subtypes is H1N1. Single hemagglutinin-neuraminidase proteins, in which both HA and NA are found in a single protein, also exist. However, these will not be used in SensUs 2021. | ||
| − | ==Mechanism of | + | ==Mechanism of infection== |
| + | [[File:Flu-infect.jpg |right|thumb|200px|An influenza virus infecting a cell.<ref name="Arti31">Sugars on Cell Surface Are Key to Flu Infections, National Institute of Health, 2008, https://www.nih.gov/news-events/nih-research-matters/sugars-cell-surface-are-key-flu-infections</ref>]] | ||
| + | |||
The involvement of the hemagglutinin and neuraminidase proteins in the infection of H1N1 is essential for the virus reproduction, and a more specific mechanism is discussed below. | The involvement of the hemagglutinin and neuraminidase proteins in the infection of H1N1 is essential for the virus reproduction, and a more specific mechanism is discussed below. | ||
| − | The hemagglutinin protein (HA) has the role of searching for the sialic acid receptors in respiratory-lining cell membranes. Upon binding of this protein and the receptor, fusion of the virus and the cell membrane is facilitated with the help of glycan proteins. The virus then enters the cell where it sheds its shell and approaches the cell’s nucleus. Using the host replication mechanisms, the virus | + | The hemagglutinin protein (HA) has the role of searching for the sialic acid receptors in respiratory-lining cell membranes. Upon binding of this protein and the receptor, fusion of the virus and the cell membrane is facilitated with the help of glycan proteins. The virus then enters the cell where it sheds its shell and approaches the cell’s nucleus. Using the host replication mechanisms, copies of the virus are made. At this step of the infection process, important viral proteins are synthesized. These newly replicated viral elements subsequently leave through the cell membrane and infect other cells. To inhibit the exit of the viral components, sialic acid receptors on the cell can bind the HA glycoproteins. This is where viral evolution/mutability can play a role in the expansion of the capabilities of the virus. |
The neuraminidase glycoprotein (N) has the role of cleaving the sialic acid receptors, allowing the exit of the viral components which then go in search of a new host. | The neuraminidase glycoprotein (N) has the role of cleaving the sialic acid receptors, allowing the exit of the viral components which then go in search of a new host. | ||
After infection is complete, the H1N1 virus triggers cell apoptosis, leading to the death of the cell and spread of the virions. | After infection is complete, the H1N1 virus triggers cell apoptosis, leading to the death of the cell and spread of the virions. | ||
| − | |||
| − | ==Medical | + | ==Medical application== |
| − | The influenza virus spreads when people with the flu cough, sneeze or talk. Infected people transfer tiny droplets to people close to them and infect them by the handover of droplets | + | The influenza virus spreads when people with the flu cough, sneeze or talk. Infected people transfer tiny droplets to people close to them and infect them by the handover of droplets. During an outbreak, it is important to be able to control and prevent the virus from spreading further, for example, by implementing control measures. Those control measures can be determined based on the reproduction number. The reproduction number is defined as: “the expected number of secondary cases produced by a single infection in a completely susceptible population”.<ref name="Arti16">Notes on R0, 2007, James Holland Jones, https://web.stanford.edu/~jhj1/teachingdocs/Jones-on-R0.pdfe</ref> A clear overview of the number of infected people is needed to calculate the reproduction number. |
| − | For disease control, it is crucial to know the infection rate in a specific region. The results may influence critical decisions such as whether to perform other diagnostic testing or to implement infection prevention and control measures for influenza | + | For disease control, it is crucial to know the infection rate in a specific region. The results may influence critical decisions such as whether to perform other diagnostic testing or to implement infection prevention and control measures for influenza. Furthermore, manpower can be in short supply during a pandemic and the speed at which someone can be tested and receive the result is of vital importance. |
| − | Biosensors that will be developed in SensUs 2021 are envisioned to be used outside the hospital in a point-of-care (POC) setting. Inside the hospital there is not a big advantage, as there are already very specific and accurate tests available for that setting | + | |
| + | Biosensors that will be developed in SensUs 2021 are envisioned to be used outside the hospital in a point-of-care (POC) setting. Inside the hospital there is not a big advantage, as there are already very specific and accurate tests available for that setting.<ref name="Arti28">Diagnosing Flu, CDC, 2020, https://www.cdc.gov/flu/symptoms/testing.htm</ref><ref name="Arti29">Information on Rapid Molecular Assays, RT-PCR, and other Molecular Assays for Diagnosis of Influenza Virus Infection, CDC, 2020, https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm</ref> Important applications are fast testing at the general practioner and at home. Therefore the test should be easy to use. The biosensors will be designed to enable a fast yes/no answer, based on measuring the concentration of hemagglutinin particles in the sample. The biosensors will not distinguish between different virus subtypes, as the subtype causing the pandemic is assumed to be known. | ||
==State of the Art== | ==State of the Art== | ||
There are various influenza tests available on the market. The most common tests are ‘rapid influenza diagnostic tests’, also called RIDTs. RIDTs provide results in a qualitative way within approximately 10-15 minutes and work by detecting the parts of the virus that stimulate an immune response.<ref name="Arti17">Rapid Influenza Diagnostic Tests, 2016, Centers for Disease Control and Prevention, https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm</ref> | There are various influenza tests available on the market. The most common tests are ‘rapid influenza diagnostic tests’, also called RIDTs. RIDTs provide results in a qualitative way within approximately 10-15 minutes and work by detecting the parts of the virus that stimulate an immune response.<ref name="Arti17">Rapid Influenza Diagnostic Tests, 2016, Centers for Disease Control and Prevention, https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm</ref> | ||
| − | The table contains examples of available | + | The table contains examples of available rapid influenza diagnostic tests that provide results in 10-15 minutes. In the SensUs Competition, a saliva based test will be developed. Moreover, SensUs strives to stimulate developement of sensors that provide results within 5-10 minutes and that are as sensitive as possible. The SensUs Competition aims to innovate the field of influenza biosensing by using saliva as a matrix, by improving the speed of the test, and by targeting a high sensitivity. |
| − | |||
| − | |||
| − | In the SensUs Competition, a saliva based test will be developed. Moreover, SensUs strives to | ||
{| class="wikitable" style="margin-bottom:0" | {| class="wikitable" style="margin-bottom:0" | ||
| Line 61: | Line 58: | ||
!Sample type | !Sample type | ||
!LOD*** (H1N1) | !LOD*** (H1N1) | ||
| − | TCID50/mL | + | TCID50/mL**** |
|- | |- | ||
|Abbott<ref name="Arti18">Abbott RIDT Product Information, Abbott, https://www.globalpointofcare.abbott/nl/product-details/binaxnow-influenza-a-and-b.html</ref> | |Abbott<ref name="Arti18">Abbott RIDT Product Information, Abbott, https://www.globalpointofcare.abbott/nl/product-details/binaxnow-influenza-a-and-b.html</ref> | ||
| Line 70: | Line 67: | ||
|90-99% | |90-99% | ||
|NPS, NS direct | |NPS, NS direct | ||
| − | | | + | |N/A |
|- | |- | ||
|Becton Dickinson & Co.<ref name="Arti19">Becton Dickinson & Co. RIDT Product Information, Becton Dickinson & Co., https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/point-of-care-testing/veritor-system</ref> | |Becton Dickinson & Co.<ref name="Arti19">Becton Dickinson & Co. RIDT Product Information, Becton Dickinson & Co., https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/point-of-care-testing/veritor-system</ref> | ||
| Line 97: | Line 94: | ||
|95-96% | |95-96% | ||
|NS, NPS, NPA, NPW direct, NP, NPA, NPW in VTM | |NS, NPS, NPA, NPW direct, NP, NPA, NPW in VTM | ||
| − | | | + | |N/A |
|- | |- | ||
|Quidel Corp<ref name="Arti21">Quidel QuickVue RIDT Product Information, Quidel, https://www.quidel.com/sites/default/files/product/documents/EF1350313EN00_1.pdf</ref> | |Quidel Corp<ref name="Arti21">Quidel QuickVue RIDT Product Information, Quidel, https://www.quidel.com/sites/default/files/product/documents/EF1350313EN00_1.pdf</ref> | ||
|QuickVue® Influenza A + B | |QuickVue® Influenza A + B | ||
|Yes | |Yes | ||
| − | |N | + | |N/A |
|81,5% | |81,5% | ||
|97,8% | |97,8% | ||
| Line 116: | Line 113: | ||
|99,4% | |99,4% | ||
|NS, NPS direct, NPA(waived), NPW(not waived) | |NS, NPS direct, NPA(waived), NPW(not waived) | ||
| − | |N | + | |N/A |
|- | |- | ||
|Remel/Thermo Fisher.<ref name="Arti23">XPECT™ RIDT Product Information, Remel/Thermo Fisher, https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FMBD%2FInstructions%2FIFU24600%2520.pdf&title=WHBlY3QgRmx1IEEgYW5kIEI=</ref> | |Remel/Thermo Fisher.<ref name="Arti23">XPECT™ RIDT Product Information, Remel/Thermo Fisher, https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FMBD%2FInstructions%2FIFU24600%2520.pdf&title=WHBlY3QgRmx1IEEgYW5kIEI=</ref> | ||
| Line 159: | Line 156: | ||
'''*'''PPA: positive percent agreement (True positive / total true positive) | '''*'''PPA: positive percent agreement (True positive / total true positive) | ||
| − | '''**''' | + | '''**'''NPA: negative percent agreement (True negative / total true negative) |
'''***'''LOD: Limit of detection | '''***'''LOD: Limit of detection | ||
| − | '''**** | + | '''****'''TCID50/mL represents the viral load at which 50% of cells are infected when a solution containing the virus is added to the cell culture.<ref name="Arti32">Limits of detection for FDA-authorized COVID-19 diagnostics, BioCentury, 2020, https://www.biocentury.com/article/304801/limits-of-detection-for-fda-authorized-covid-19-diagnostics</ref> Detection limits are often expressed in TCID50/mL. Copies/TCID50 for H1N1 is 2381 copies/TCID50 so 1 TCID50/mL represents 2381 copies per mL. It is a very rough method to determine concentration and it has a standard deviation of about 1048 copies/TCID50.<ref name="Arti33">Analytical Sensitivity Comparison between Singleplex Real-Time PCR and a Multiplex PCR Platform for Detecting Respiratory Viruses, PLoS One, 2015,Parker J, Fowler N, Walmsley ML, Schmidt T, Scharrer J, Kowaleski J, Grimes T, Hoyos S, Chen J., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4646456/</ref></sub></div> |
| + | |||
| + | CLIA waived tests are tests cleared for POC setting. These are simple tests with a relatively low risk for an incorrect result. Tests are scored by seven criteria on a scale of one to three, where a score of 1 indicates the lowest level of complexity. The tests may be called CLIA waived, if the score is low enough. The seven criteria are:<ref name="Arti27">Categorization Criteria CLIA test, FDA, 2020, Inc., https://www.fda.gov/node/365445#scorecard</ref> | ||
| + | |||
| + | 1. Knowledge: scientific and technical knowledge required to use the test. | ||
| + | |||
| + | 2. Training and experience: training needed to perform the test. | ||
| + | |||
| + | 3. Reagents and materials preparation: stability of materials and material preparation. | ||
| + | |||
| + | 4. Characteristics of operational steps: complexity of operational steps. | ||
| + | |||
| + | 5. Calibration, quality control and proficiently testing materials: can be stable or labile. | ||
| + | |||
| + | 6. Test system troubleshooting and equipment maintenance: the easiness and frequency of the troubleshooting and equipment maintenance. | ||
| + | |||
| + | 7. Interpretation and judgement: required to perform analytic processes and resolution of problems. | ||
| + | |||
| + | An alternative type of testing is with a ‘molecular assay’, which is more accurate than RITDs, because it detects viral RNA or nucleic acids in respiratory specimens. This type of testing includes rapid molecular assays, RT-PCR and other nucleic acid amplification tests. Molecular tests are mostly used in hospitals, some are able to detect both influenza A and B. Others can identify different subtypes. The ‘molecular assays’ are more accurate and sensitive than RIDTs, but the time-to-result may be several hours.<ref name="Arti28">Diagnosing Flu, CDC, 2020, https://www.cdc.gov/flu/symptoms/testing.htm</ref><ref name="Arti29">Information on Rapid Molecular Assays, RT-PCR, and other Molecular Assays for Diagnosis of Influenza Virus Infection, CDC, 2020, https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm</ref> | ||
==Safety== | ==Safety== | ||
| + | When working with the influenza virus, influenza infection in humans can occur following a laboratory accident. For safety reasons, unbound hemagglutinin particles will be used in SensUs 2021, as a substitute for infectious influenza virus particles. | ||
| − | + | ===Lab protocols=== | |
| + | The use of safety equipment combined with good practices is fundamental to laboratory safety and in helping to reduce the risks involved in dealing with biosafety hazards. Therefore, it is important that you consult with your local biosafety officer and comply to the safety rules of your own organization. | ||
| − | + | According to Article 4.84 of the Working Conditions Decree in The Netherlands, inactivated virus particles can be classified as category 1 of the biological agents<ref name="Arti30">1Short manual for the ML-I, ML-II laboratory(gmo-labs) and/or laboratory for working with biological agents / human materials, University of Twente, 2018,https://www.utwente.nl/.uc/f4fcab199010209618400ac90d20284e61d13c337a78900/short-manual-gmo.pdf</ref>, as they are unlikely to cause disease in humans. Therefore, the biosafety guidelines as defined in the ML-1 (minimum containment level) have to be followed. It is stipulated in the guidelines that general laboratory protocols such as the usage of barrier protection (lab coats, gloves, and face protection) when handling the samples are to be followed. The protection barriers are not always necessary from a microbiological perspective. However, it is compulsory when handling disinfectants or solvents. | |
| − | |||
| − | According to Article 4.84 of the Working Conditions Decree, | ||
| − | Therefore, | ||
== References == | == References == | ||
<references /> | <references /> | ||
Latest revision as of 14:20, 29 January 2021
Contents
General information
The theme of SensUs 2021 is acute respiratory viruses. The current Covid-19 pandemic has made it apparent that large virus outbreaks can cause immense harm to human health and can disrupt society as a whole. One of the most common respiratory viruses is influenza A. Therefore, the influenza virus serves as an interesting research topic for this year’s Competition.
The influenza A virus is known to cause influenza in birds and some mammals, like humans. Several subtypes of the influenza A virus have been isolated from wild birds. Occasionally, viruses can be transmitted from wild birds to domestic animals, like chickens or pigs. This may give rise to human influenza.[2]
Influenza A viruses are negative-sense, single-stranded RNA viruses. Subtypes of influenza A are characterized by proteins on the outermost membrane of the virus, called hemagglutinin (H or HA) and neuraminidase (N or NA). H and N are the antigens of the virus, and play an important role in the interaction between the host’s immunological response and the virus. Recently, researchers have reported the discovery of an antibody that is generally effective against all subtypes of the influenza A virus.[3] Hemagglutinin will serve as this year's biomarker. In infected patients, bound hemagglutinin indicates the presence of the influenza virus. In the Testing Event, unbound hemagglutinin proteins will be used to mimic the immunologic response of the virus.
The subtype which will be used in SensUs 2021 is H1N1. Historically, H1N1 has been responsible for most deaths due to influenza. It is a popular influenza strain for research purposes.[4][5][6] Due to its popularity among researchers, antigens and antibodies are commercially available, making H1N1 suitable as a target for the SensUs Competition. Influenza A vaccines for humans have been developed. New versions of the vaccines are developed twice per year for use all over the world, which is necessary due to rapid mutations of the influenza virus. Every year during flu season, a large part of the population is vaccinated in order to protect individuals against the virus. However, due to unforeseen mutations of the virus, it might be possible that in a certain year a vaccine will prove ineffective. In that case large portions of the population would be at risk and a pandemic could occur. The probability of a major influenza A pandemic is estimated to be around 0.5-1% each year.[7]
History of influenza (A)
A lack of data up until 1500 AC complicates the research on influenza before that period.[8] Possibly the first influenza pandemic occurred around 6000 BC in China. The symptoms of human influenza seem to have been clearly described by Hippocrates, roughly 2,400 years ago[9] Although the virus seems to have caused epidemics throughout human history, historical data on influenza is difficult to interpret, due to the fact that symptoms of influenza are similar to those found in other respiratory diseases, like respiratory syncytial virus (RSV).
The most infamous and lethal outbreak was the 1918 flu pandemic, which lasted until 1920. The number of deaths is unknown, but estimates range from 17 to 100 million people.[10]. This pandemic has been described as "the greatest medical holocaust in history"[11] and may have killed as many people as the plague (Black Death). This huge death toll was caused by an extremely high infection rate of up to 50% and the severity of the symptoms, suspected to be caused by cytokine storms in which the innate immune system causes an uncontrolled and excessive release of pro-inflammatory signaling molecules called cytokines. One of the most recent outbreaks of influenza was the 2009 Swine Flu. Similar to the Spanish Flu, it was also of the subtype H1N1. The death toll of the 2009 pandemic is estimated to be around 150,000 to 575,000.[12]
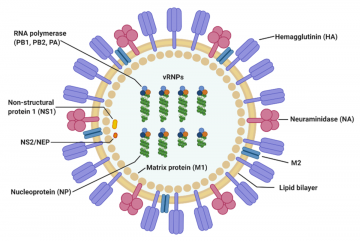
Structure of the influenza virus
The virus particle (virion) is 80–120 nanometers in diameter.[13] The virion shape can be spherical, elliptical, or even filamentous with a length of tens of micrometers[13]. The virion is made up of a viral envelope containing two main types of proteins, wrapped around a central core. The two large proteins found on the outside of viral particles are hemagglutinin (HA) and neuraminidase (NA). HA is a protein that mediates binding of the virion to target cells and entry of the viral genome into the target cell, and therefore plays an important role in infecting healthy cells. NA is involved in releasing the progeny viruses once a cell has been infected and has started producing the virus itself.[14]
These two proteins are a target of interest for antiviral drugs.[15] Furthermore, they are also the antigen proteins to which a host antibodies can bind and trigger an immune response. Influenza type A viruses are categorized into different subtypes, or strains, based on which type of these two proteins is present on the surface of the virion. Currently, there are 16 subtypes of HA and 9 subtypes of NA known to exist. The most prevalent form of the different subtypes is H1N1. Single hemagglutinin-neuraminidase proteins, in which both HA and NA are found in a single protein, also exist. However, these will not be used in SensUs 2021.
Mechanism of infection

The involvement of the hemagglutinin and neuraminidase proteins in the infection of H1N1 is essential for the virus reproduction, and a more specific mechanism is discussed below. The hemagglutinin protein (HA) has the role of searching for the sialic acid receptors in respiratory-lining cell membranes. Upon binding of this protein and the receptor, fusion of the virus and the cell membrane is facilitated with the help of glycan proteins. The virus then enters the cell where it sheds its shell and approaches the cell’s nucleus. Using the host replication mechanisms, copies of the virus are made. At this step of the infection process, important viral proteins are synthesized. These newly replicated viral elements subsequently leave through the cell membrane and infect other cells. To inhibit the exit of the viral components, sialic acid receptors on the cell can bind the HA glycoproteins. This is where viral evolution/mutability can play a role in the expansion of the capabilities of the virus. The neuraminidase glycoprotein (N) has the role of cleaving the sialic acid receptors, allowing the exit of the viral components which then go in search of a new host. After infection is complete, the H1N1 virus triggers cell apoptosis, leading to the death of the cell and spread of the virions.
Medical application
The influenza virus spreads when people with the flu cough, sneeze or talk. Infected people transfer tiny droplets to people close to them and infect them by the handover of droplets. During an outbreak, it is important to be able to control and prevent the virus from spreading further, for example, by implementing control measures. Those control measures can be determined based on the reproduction number. The reproduction number is defined as: “the expected number of secondary cases produced by a single infection in a completely susceptible population”.[17] A clear overview of the number of infected people is needed to calculate the reproduction number.
For disease control, it is crucial to know the infection rate in a specific region. The results may influence critical decisions such as whether to perform other diagnostic testing or to implement infection prevention and control measures for influenza. Furthermore, manpower can be in short supply during a pandemic and the speed at which someone can be tested and receive the result is of vital importance.
Biosensors that will be developed in SensUs 2021 are envisioned to be used outside the hospital in a point-of-care (POC) setting. Inside the hospital there is not a big advantage, as there are already very specific and accurate tests available for that setting.[18][19] Important applications are fast testing at the general practioner and at home. Therefore the test should be easy to use. The biosensors will be designed to enable a fast yes/no answer, based on measuring the concentration of hemagglutinin particles in the sample. The biosensors will not distinguish between different virus subtypes, as the subtype causing the pandemic is assumed to be known.
State of the Art
There are various influenza tests available on the market. The most common tests are ‘rapid influenza diagnostic tests’, also called RIDTs. RIDTs provide results in a qualitative way within approximately 10-15 minutes and work by detecting the parts of the virus that stimulate an immune response.[20] The table contains examples of available rapid influenza diagnostic tests that provide results in 10-15 minutes. In the SensUs Competition, a saliva based test will be developed. Moreover, SensUs strives to stimulate developement of sensors that provide results within 5-10 minutes and that are as sensitive as possible. The SensUs Competition aims to innovate the field of influenza biosensing by using saliva as a matrix, by improving the speed of the test, and by targeting a high sensitivity.
| Manufacturer | Product | CLIA Waived | Platform | Sensitivity
(PPA*) |
Specificity
(NPA**) |
Sample type | LOD*** (H1N1)
TCID50/mL**** |
|---|---|---|---|---|---|---|---|
| Abbott[21] | Binax Now Influenza A & B Card 2 | Yes | DIGIVAL | 70-89% | 90-99% | NPS, NS direct | N/A |
| Becton Dickinson & Co.[22] | BD Veritor™ Flu A + B | Yes | BD veritor Reader | 82% | 98% | NPS, NS direct | 3.3*102-5.0*103 |
| Becton Dickinson & Co.[22] | BD Veritor™ Flu A + B | Yes | BD veritor plus Analyzer | 83,6% | 97,5% | NPS, NS direct | 3.3*102-5.0*103 |
| Quidel Corp.[23] | Sofia® Influenza A + B FIA | Yes | Sofia(2) FIA Analyzer | 90-99% | 95-96% | NS, NPS, NPA, NPW direct, NP, NPA, NPW in VTM | N/A |
| Quidel Corp[24] | QuickVue® Influenza A + B | Yes | N/A | 81,5% | 97,8% | NS, NPS, NPA, NPW direct, NP, NPA, NPW in VTM | 1.63*103-4.4*103 |
| Princeton BioMeditech Corp.[25] | BioSign® Flu A & B
LABSCO |
Yes | N/A | 89,2% | 99,4% | NS, NPS direct, NPA(waived), NPW(not waived) | N/A |
| Remel/Thermo Fisher.[26] | XPECT™ Flu A & B
LABSCO |
No | N/A | 89-100% | 100% | NW, NS | 1.63*103-4.41*103 |
| Sekisui Diagnostics, [27] | Acucy Influenza A&B Test | Yes | The Acucy System | 96,4% | 96,0% | NPS, NS direct | 1.4*101 |
| Sekisui Diagnostics, [28] | OSOM Ultra Plus Flu A&B Test | Yes | N/A | 90,2-92,2% | 75,1-85,7% | NPS, NPA, NPW, NS | 1.05*102 |
| Access Bio, Inc., [29] | CareStart Flu A&B Plus | No | N/A | 79,9% | 98,4% | NPS | 5.0*103-9.6*103
|
*PPA: positive percent agreement (True positive / total true positive)
**NPA: negative percent agreement (True negative / total true negative)
***LOD: Limit of detection
****TCID50/mL represents the viral load at which 50% of cells are infected when a solution containing the virus is added to the cell culture.[30] Detection limits are often expressed in TCID50/mL. Copies/TCID50 for H1N1 is 2381 copies/TCID50 so 1 TCID50/mL represents 2381 copies per mL. It is a very rough method to determine concentration and it has a standard deviation of about 1048 copies/TCID50.[31]CLIA waived tests are tests cleared for POC setting. These are simple tests with a relatively low risk for an incorrect result. Tests are scored by seven criteria on a scale of one to three, where a score of 1 indicates the lowest level of complexity. The tests may be called CLIA waived, if the score is low enough. The seven criteria are:[32]
1. Knowledge: scientific and technical knowledge required to use the test.
2. Training and experience: training needed to perform the test.
3. Reagents and materials preparation: stability of materials and material preparation.
4. Characteristics of operational steps: complexity of operational steps.
5. Calibration, quality control and proficiently testing materials: can be stable or labile.
6. Test system troubleshooting and equipment maintenance: the easiness and frequency of the troubleshooting and equipment maintenance.
7. Interpretation and judgement: required to perform analytic processes and resolution of problems.
An alternative type of testing is with a ‘molecular assay’, which is more accurate than RITDs, because it detects viral RNA or nucleic acids in respiratory specimens. This type of testing includes rapid molecular assays, RT-PCR and other nucleic acid amplification tests. Molecular tests are mostly used in hospitals, some are able to detect both influenza A and B. Others can identify different subtypes. The ‘molecular assays’ are more accurate and sensitive than RIDTs, but the time-to-result may be several hours.[18][19]
Safety
When working with the influenza virus, influenza infection in humans can occur following a laboratory accident. For safety reasons, unbound hemagglutinin particles will be used in SensUs 2021, as a substitute for infectious influenza virus particles.
Lab protocols
The use of safety equipment combined with good practices is fundamental to laboratory safety and in helping to reduce the risks involved in dealing with biosafety hazards. Therefore, it is important that you consult with your local biosafety officer and comply to the safety rules of your own organization.
According to Article 4.84 of the Working Conditions Decree in The Netherlands, inactivated virus particles can be classified as category 1 of the biological agents[33], as they are unlikely to cause disease in humans. Therefore, the biosafety guidelines as defined in the ML-1 (minimum containment level) have to be followed. It is stipulated in the guidelines that general laboratory protocols such as the usage of barrier protection (lab coats, gloves, and face protection) when handling the samples are to be followed. The protection barriers are not always necessary from a microbiological perspective. However, it is compulsory when handling disinfectants or solvents.
References
- ↑ Host Protective Immune Responses against Influenza A Virus Infection, MDPI, 2020, Hi Eun Jung, Heung Kyu Lee, https://www.mdpi.com/1999-4915/12/5/504/htm
- ↑ Transmission of Avian Influenza A Viruses Between Animals and People, CDC, 2015, https://www.cdc.gov/flu/avianflu/virus-transmission.htm
- ↑ Super antibody' fights off flu, BBC, 2011, James Gallagher, https://www.bbc.com/news/health-14324901
- ↑ Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection, Research Gate, 2020, Sun, Honglei & Xiao, Yihong & Liu, Jiyu & Wang, Dayan & Li, Fangtao & Wang, Chenxi & Li, Chong & Zhu, Junda & Song, Jingwei & Sun, Haoran & Zhimin, Jiang & Liu, Litao & Zhang, Xin & Wei, Kai & Dongjun, Hou & Pu, Juan & Sun, Yipeng & Tong, Qi & Bi, Yuhai & Liu, Jinhua https://www.researchgate.net/publication/342555087_Prevalent_Eurasian_avian-like_H1N1_swine_influenza_virus_with_2009_pandemic_viral_genes_facilitating_human_infection
- ↑ Comparison of Hospitalized Patients With ARDS Caused by COVID-19 and H1N1, CHEST, Xiao Tang, Rong-Hui Du, Rui Wang, Tan-Ze Cao, Lu-Lu Guan, Cheng-Qing Yang, Qi Zhu, Ming Hu, Xu-Yan Li, Ying Li, Li-Rong Liang, Zhao-Hui Tong, Bing Sun, Peng Peng, Huan-Zhong Shi, 2020, https://www.sciencedirect.com/science/article/pii/S0012369220305584
- ↑ Landscape of coordinated immune responses to H1N1 challenge in humans, Journal of Clinical Investigation, 2020, Zainab Rahil, Rebecca Leylek, Christian M. Schürch, Han Chen, Zach Bjornson-Hooper, Shannon R. Christensen, Pier Federico Gherardini, Salil S. Bhate, Matthew H. Spitzer, Gabriela K. Fragiadakis, Nilanjan Mukherjee, Nelson Kim, Sizun Jiang, Jennifer Yo, Brice Gaudilliere, Melton Affrime, Bonnie Bock, Scott E. Hensley, Juliana Idoyaga, Nima Aghaeepour, Kenneth Kim, Garry P. Nolan, David R. McIlwain https://www.sciencedirect.com/science/article/pii/S0012369220305584
- ↑ Pandemic risk: how large are the expected losses?, WHO, 2017, Victoria Y Fan, Dean T Jamisonb & Lawrence H Summers, https://www.who.int/bulletin/volumes/96/2/17-199588.pdf
- ↑ Internet‐Based Intelligence in Public Health Emergencies, NATO Science for Peace and Security Series - E: Human and Societal Dynamics, 2013 Mordini E., Green M., https://www.iospress.nl/book/internet%E2%80%90based-intelligence-in-public-health-emergencies/
- ↑ 2,500-year Evolution of the Term Epidemic, Emerging infectious diseases, 2006, Martin PM, Martin-Granel E, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3373038/
- ↑ Reassessing the Global Mortality Burden of the 1918 Influenza Pandemic, American journal of epidemiology, 2018, Spreeuwenberg P, Kroneman M, Paget J, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7314216/
- ↑ Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission , Pathogens, 2016, Saunders-Hastings PR, Krewski D, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5198166/
- ↑ 2009 H1N1 Pandemic (H1N1pdm09 virus), centers for disease control and prevention, 2010, https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html
- ↑ 13.0 13.1 Native Morphology of Influenza Virions, Frontiers in microbiology, 2011, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249889/
- ↑ Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase, Virology journal , 2013, Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3842836/
- ↑ Recent Strategies in the Search for New Anti-Influenza Therapies, Current Drug Targets , 2003, J.C. Wilson, M. von Itzstein, https://www.eurekaselect.com/63785/article
- ↑ Sugars on Cell Surface Are Key to Flu Infections, National Institute of Health, 2008, https://www.nih.gov/news-events/nih-research-matters/sugars-cell-surface-are-key-flu-infections
- ↑ Notes on R0, 2007, James Holland Jones, https://web.stanford.edu/~jhj1/teachingdocs/Jones-on-R0.pdfe
- ↑ 18.0 18.1 Diagnosing Flu, CDC, 2020, https://www.cdc.gov/flu/symptoms/testing.htm
- ↑ 19.0 19.1 Information on Rapid Molecular Assays, RT-PCR, and other Molecular Assays for Diagnosis of Influenza Virus Infection, CDC, 2020, https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm
- ↑ Rapid Influenza Diagnostic Tests, 2016, Centers for Disease Control and Prevention, https://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm
- ↑ Abbott RIDT Product Information, Abbott, https://www.globalpointofcare.abbott/nl/product-details/binaxnow-influenza-a-and-b.html
- ↑ 22.0 22.1 Becton Dickinson & Co. RIDT Product Information, Becton Dickinson & Co., https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/point-of-care-testing/veritor-system
- ↑ Quidel RIDT Product Information, Quidel, https://www.quidel.com/immunoassays/rapid-influenza-tests/sofia-influenza-fia
- ↑ Quidel QuickVue RIDT Product Information, Quidel, https://www.quidel.com/sites/default/files/product/documents/EF1350313EN00_1.pdf
- ↑ BioSing RIDT Product Information, Princeton BioMeditech Corp., http://punchout.medline.com/product/BioSign-Rapid-Flu-AB-Antigen-Panel-Test-by-Princeton-BioMeditech/Influenza-Testing/Z05-PF176440?question=&index=P11&indexCount=11#mrkSpec
- ↑ XPECT™ RIDT Product Information, Remel/Thermo Fisher, https://www.thermofisher.com/document-connect/document-connect.html?url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FMBD%2FInstructions%2FIFU24600%2520.pdf&title=WHBlY3QgRmx1IEEgYW5kIEI=
- ↑ Acucy Influenza A&B Test Product Information, Sekisui Diagnostics, https://www.sekisuidiagnostics.com/wp-content/uploads/2019/08/Acucy-Influenza-AB-Package-Insert.pdf
- ↑ OSOM Ultra Plus Flu A&B Test Product Information, Sekisui Diagnostics, https://www.sekisuidiagnostics.com/wp-content/uploads/2019/04/OSOM_P-52631-D__web.pdf
- ↑ CareStart Flu A&B Plus Product Information, Access Bio, Inc., https://www.accessdata.fda.gov/cdrh_docs/pdf19/K191514.pdf
- ↑ Limits of detection for FDA-authorized COVID-19 diagnostics, BioCentury, 2020, https://www.biocentury.com/article/304801/limits-of-detection-for-fda-authorized-covid-19-diagnostics
- ↑ Analytical Sensitivity Comparison between Singleplex Real-Time PCR and a Multiplex PCR Platform for Detecting Respiratory Viruses, PLoS One, 2015,Parker J, Fowler N, Walmsley ML, Schmidt T, Scharrer J, Kowaleski J, Grimes T, Hoyos S, Chen J., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4646456/
- ↑ Categorization Criteria CLIA test, FDA, 2020, Inc., https://www.fda.gov/node/365445#scorecard
- ↑ 1Short manual for the ML-I, ML-II laboratory(gmo-labs) and/or laboratory for working with biological agents / human materials, University of Twente, 2018,https://www.utwente.nl/.uc/f4fcab199010209618400ac90d20284e61d13c337a78900/short-manual-gmo.pdf